Hypoxia Inducible FactorsHypoxia-inducible factors (HIFs) are transcription factors that respond to decreased oxygen level in the cellular environment. Various isoforms and splice variants have been identified and their activities vary according with their specific expression patterns and structural domain composition.
|
||||||||||||||||||||
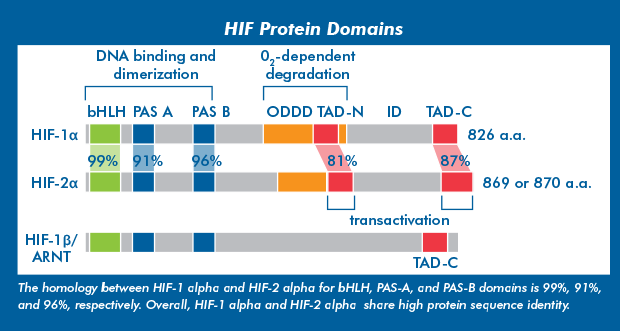
HIF-1 and -2, alphaHypoxia inducible factor 1 (HIF-1) was first identified in the early 1990s and molecularly characterized as a heterodimer consisting of an alpha (oxygen-sensitive) and beta (oxygen-insensitive) subunits. Two additional isoforms, HIF-2 alpha and HIF-3 alpha, were subsequently identified. HIF-1 alpha and HIF-2 alpha are generally differentially expressed, but share several transcriptional targets with an overall positive response to hypoxia. Lastly, HIF-3 alpha is less well studied but is known to have both positive and negative effects on hypoxia responses. |
||||||||||||||||||||
|
||||||||||||||||||||
|
HIF-beta subunits are constitutively expressed, independently of oxygen levels, and present in the nucleus. HIF-alpha subunits are expressed constitutively, and predominantly regulated post-translationally. Recently, epigenetic mechanisms have been shown to regulate the transcription of HIF-1 alpha and HIF-2 alpha in response to hypoxia. |
||||||||||||||||||||
The HIF-alpha subunit isoforms form heterodimers with HIF-1 beta (aryl hydrocarbon receptor nuclear translocators- ARNT) or HIF-2 beta (ARNT2), and as a complex regulate gene expression. |
||||||||||||||||||||
HIF-3 alpha |
||||||||||||||||||||
|
Alternative splicing of HIF-3 alpha mRNA and multiple transcriptional start sites within the HIF-3 alpha gene result in several mRNA products and protein variants. Some of these products completely lack transactivation domains and are unable to regulate gene expression. HIF-3 alpha subunits without TAD, also known as PAS domain proteins (IPAS), act as inhibitors of both HIF-1 and HIF-2 activity. HIF-3 isoforms with an TAD-N sequence are able to regulate gene expression under hypoxia. Because all HIF-3 isoforms contain bHLH and PAS domains, variants form heterodimers with HIF- beta subunits. |
||||||||||||||||||||

|
||||||||||||||||||||
|
HIF-3 alpha transcripts are broadly expressed in human tissues and organs, with some variants present at higher levels at embryonic stages. Many HIF-3 alpha variants contain an ODDD sequence, and therefore protein stabilization under hypoxia is expected. However, HIF-3 alpha is known to be regulated via mechanism that target its transcription, translation and nuclear localization. |
||||||||||||||||||||
|
||||||||||||||||||||
Post-translational Regulation of HIF-alpha subunitsHIF-alpha subunits are regulated by four oxygen-sensing enzymes including prolyl hydroxylases (PHD1, PHD2 and PHD3) and factor inhibiting HIF (FIH).
Major Enzymes Regulating HIF-1 alpha |
||||||||||||||||||||

|
||||||||||||||||||||
|
|
||||||||||||||||||||